Automatic population of Report times
ARISj does NOT automatically calculate the follow-up number for an initial/follow-up report. This information is collected on Informed Authority screen and is currently entered manually by the business user. After the suggested functionality implementation the users would no longer be required to enter the follow-up information manually. As long as the acknowledgement is imported using ESM, this calculation will be performed.
Assumptions
This calculation will be done at ESM when the case is exported
This will be applicable only for Japanese transactions
This automatic calculation will be applied upon export of a case
Only J2(M2) Calculation will be performed when the "Resubmission" field is checked for the Informed Authority record for the version of the case.
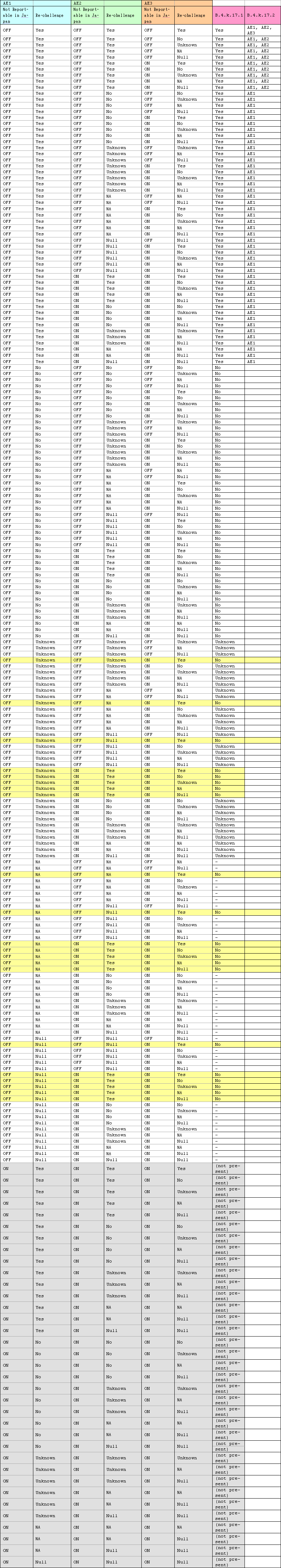
Calculation Logic
The J.2, M.2 and J.5 fields are to be automatically calculated by ESMServer and update to ARISj database base on the following logic.
These tags should be calculated based on the A.1.6 (Message Transmission Acknowledgement Code) and B.1.8 (Report Acknowledgement code) fields of the acknowledgement of the previous transmission.
For the initial transmission of the case, the J, 5, M.2 and J.2 fields should have the value 1. This should be updated back in the AER into the corresponding informed authority record.
It is assumed that when a case is submitted, the user creates a new informed authority record with the report type and with re-submission filed setting to the right value.
ARISj should copy the J.2, M.2 and J.5 values from the previous informed authority record when a new record is created for the same authority and report type if there is a previous informed authority record. For Japanese multi reporting drug also will be considered while selecting the previous informed authority record and while copy it will copy the safetyreporid from the previous record.
For each of the successive submissions, the latest authority record should be updated with the calculated value depending on the logic below. Please note that the calculation should be done based on the Acknowledgement received for the previous transmission
The ICSR file should have the newly calculated values for J.2,M.2 and J.5
|
ACK from previous report |
Current ICSR+J file |
|
||
|
A.1.6 |
B.1.8 |
J2(M2) |
J5 |
Data accepted at MHLW? |
|
01 |
01 |
+1 |
+1 |
Yes |
|
01 |
02 |
+1 |
+1 |
Yes |
|
02 |
01 |
Invalid combination |
No |
|
|
02 |
02 |
+1 |
Same as previous report |
No |
|
03 |
01 |
Invalid combination |
No |
|
|
03 |
02 |
+1 |
Same as previous report |
No |
|
No ACK in case of parse error, file name error, etc. MHLW will send the company an email to notify. Resubmission of the case required. |
+1 |
Same as previous report |
No |
|
The value of the "Resubmission" field should be considered in automatic calculation logic for the J.2, M.2 and J.5 fields. When the value of the "Resubmission" field is considered, the logic is as the following table:
|
ACK from previous report |
"Resubmission" field |
Data accepted at MHLW? |
Current ICSR+J file |
||
|
A.1.6 |
B.1.8 |
J2(M2) |
J5 |
||
|
01 |
01 |
Uncheck |
Yes |
+1 |
+1 |
|
01 |
01 |
Check |
Yes But re-submission is required. |
+1 |
Same as previous report |
|
01 |
02 |
Uncheck |
Yes |
+1 |
+1 |
|
01 |
02 |
Check |
Yes But re-submission is required. |
+1 |
Same as previous report |
|
02 |
01 |
Regardless of the value for "Resubmission" field |
No |
Invalid combination |
|
|
02 |
02 |
Regardless of the value for "Resubmission" field |
No |
+1 |
Same as previous report |
|
03 |
01 |
Regardless of the value for "Resubmission" field |
No |
Invalid combination |
|
|
03 |
02 |
Regardless of the value for "Resubmission" field |
No |
+1 |
Same as previous report |
|
No ACK in case of parse error, file name error, etc. MHLW will send the company an email to notify. Resubmission of the case required. |
Check |
No |
+1 |
Same as previous report |
|
The system will not consider the Code broken or other fields related to blinding/unblinding in ARIS schema.
The system will consider the 1 ('Blinded') or 2 ("UnBlinded") value set in the DISTRIBUTION_OF_AER_INFO.BLINDED_REPORT column and would export the blinded/unblinded records along with normal records accordingly.
Scenarios during export:
|
SNO |
DISTRIBUTION_OF_AER_INFO (BLINDING_REPORT ) COLUMN |
OUTPUT IN ICSR |
|
1 |
1(YES ) |
NORMAL + BLINDED RECORD |
|
2 |
2(NO) |
NORMAL + UN-BLINDED RECORD |
Exception :The below mentioned third and fourth scenarios would be an exception in the current release and may not work as ARISg should be corrected to update the correct values in the DISTRIBUTION_OF_AER_INFO table.
|
SNO |
ADMIN PROTOCOL (CODE_BROKEN) |
DISTRIBUTION_OF_AER _INFO (BLINDING_REPORT ) COLUMN |
Expected OUTPUT IN ICSR |
EXCEPTIONS |
|
1 |
NO |
YES |
NORMAL + BLINDED RECORD |
|
|
2 |
NO |
NO |
NORMAL + UN-BLINDED RECORD |
|
|
3 |
YES / NA / NULL |
What Ever Specified |
NORMAL + UN-BLINDED RECORD |
This scenario will get failed, since ARISg should consider the code broken fields to override Format details set in Distribution conditions which is currently not handled. |
|
4 |
- |
NULL |
ALL Normal records |
In the current system Null will not be present in any scenario how ever if this exists then All the Normal records should be exported. But as discussed, this scenario will be a Limitation in the Current release. |
All blinded records will be exported irrespective of the "Not relevant" flag value. Unblinded records will be considered for "Not relevant" flag. When the NOT_RELEVANT Flag is checked, then that Un-blinded record will not be exported. And also the Normal records marked as "Not relevant" in ARISg product screen will not be exported.
AUTO_RANK: (The sequence in which records to be exported):
The system will be enhanced to consider the auto rank for blinded records also. The system will export the drug details in the following order for blinded and unblinded reports. That is, the ordering will be based on AER_PRODUCT.AUTO_RANK_BLIND column.
The drug blocks for Normal+blinded records will be exported in the same order in which they are displayed in the Standard ARISg product screens. That is, the ordering will be based on AER_PRODUCT.AUTO_RANK_BLIND column.
For an Unblinded Report the ordering of drug blocks in ICSR will be, first all the Normal records will be exported and then the Unblinded Records will be exported. That is, the ordering will be based on AER_PRODUCT.AUTO_RANK_BLIND column.